Kou-San Ju
Contact Information
Associate Professor of Microbiology and Medicinal Chemistry & Pharmacognosy
Areas of Expertise
- Natural products
- Microbial metabolism
- Biocatalysis
Education
- Ph.D., University of California at Davis, 2009
- Postdoctoral, University of Illinois at Urbana-Champaign, 2010-2016
Research Interests
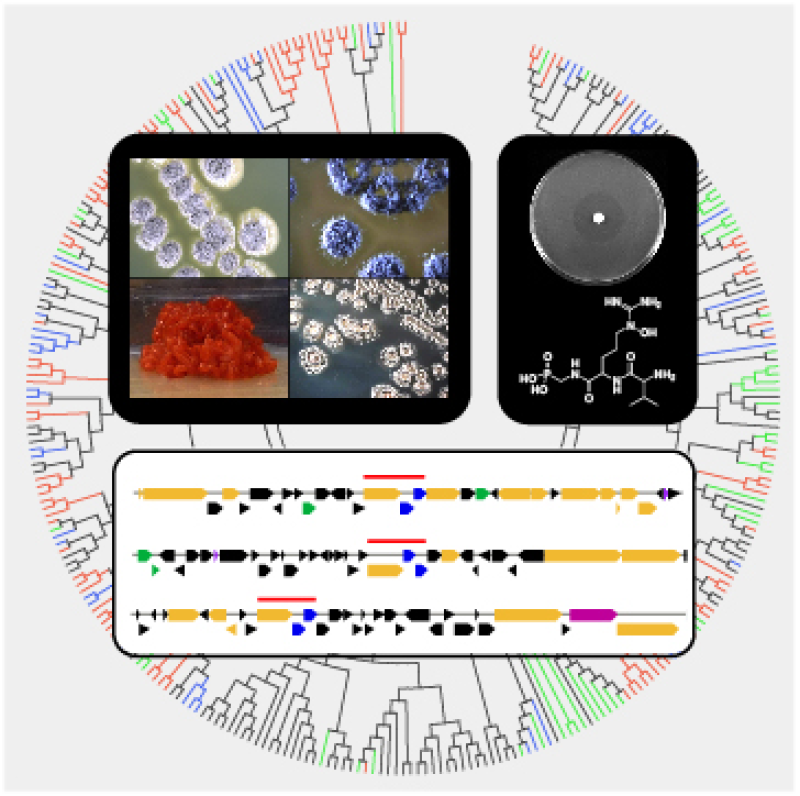
Our interdisciplinary research program is inspired by the metabolic diversity of microorganisms and the vast array of compounds they produce. Working at the interface of chemistry and biology, we combine approaches in chemistry, biochemistry, bioinformatics, genetics, and systems biology to discover new natural products, identify bioactivity and mode of action, and to decipher the metabolic basis of their biosynthesis. Ultimately, we to seek to translate insights gained from our investigations into solutions for modern day challenges facing human health and the environment. These include new antibiotics to counter drug-resistant pathogens, novel herbicides and biocontrol agents to improve pest management and food security, and engineered biocatalysts to facilitate chemical production by green chemistry and industrial biotechnology.
Genome Mining for Natural Products
We have developed a genomics-driven platform for the discovery of natural products from actinobacteria, an important source of antibiotics, pharmaceuticals and agrochemicals. In contrast to traditional bioactivity guided screening, we use low-cost, high-throughput, sequencing technologies to first classify the biosynthetic potential of strains. Combining the systematic analysis of encoded natural product gene clusters with chemospecific detection of molecules and metabolomics, an emergent path to new compounds is paved that simultaneously bypasses the “re-discovery problem” of classical discovery. Most recently, we have used this strategy to mine the genomes of 10,000 actinomycetes for phosphonic and phosphinic acids, discovering a multitude of new compounds and unexplored pathways for this understudied, but commercially proven, class of natural products.
Genome mining has also revealed that the reservoir of new compounds from actinomycetes remains vast and deep – approximately 90% of natural product gene clusters encode for unknown molecules. To harness the potential of this biosynthetic diversity, we have initiated discovery campaigns for other classes of natural products.
Bioactivity of Antibiotics
We are studying the bioactivity of newly isolated phosphonic and phosphinic acid natural products, many of which show promising antimicrobial antibiotic properties. We are particularly interested in understanding the physiological responses triggered upon natural product exposure, deducing mode of action, and identifying immunity factors. Investigations into the scope and nature of their bioactivities will provide insight into their function, feasibility as lead compounds, and potential application in other areas of biotechnology.
Deciphering Biosynthesis
We are characterizing strains that produce phosphonic acids and other natural products with unusual functional groups to understand the nature of their biosynthesis. To elucidate pathways, we construct gene-deletion mutants, analyze accumulated intermediates and products of cross-feeding experiments, and reconstitute biochemical reactions in vitro. Additionally, we seek to understand the molecular determinants of catalysis and the evolutionary origins for key biosynthetic enzymes. These results will facilitate the creation of natural product derivatives through combinatorial biosynthesis, the development of new biocatalysis agents, and inform additional markers for genome mining.
Chemical Ecology
The production, distribution, and effects of natural product molecules are largely unclear for strains when present in their natural environment. To develop a comprehensive understanding of the potential roles natural products have on the structure, function, and dynamics of microbiomes, we are examining the taxonomic and ecological distribution of biosynthetic pathways, environmental cues that trigger natural product biosynthesis, and the physiology of producing organisms. These studies will provide clues into the native function of natural products and contribute to our greater understanding of how microbes shape ecosystems.
Membership
Assistant Professor, Division of Medicinal Chemistry & Pharmacognosy (Joint Appointment)
Member, Infectious Disease Institute
Member, Center for Applied Plant Sciences
Member, Ohio State Biochemistry Graduate Program
Member, Molecular and Cellular Developmental Biology Graduate Program
Recent Publications
Goettge, M.N., J.P. Cioni, K.S. Ju, K. Pallitsch, and W.W. Metcalf. 2018. Biochemical characterization of a novel class of flavin-dependent, oxime-formining N-oxidases involved in the biosynthesis of phosphyocystoximic acids. J. Biol. Chem. 293(18):6859-6868. PMID: 29540479
Parkinson, E.I., J.H. Tryon, A.W. Goering, K.S. Ju, R.A. McClure, J.D. Kemball, S. Zhukovsky, R.J. Thomson, N.L. Kelleher, W.W. Metcalf. 2018. Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem. Biol. 13(4):1029-1037. PMID: 29510029
Born, D., E. Ulrich, K.S. Ju, S. Peck, W.A. van der Donk, and C.L. Drennan. 2017. Structural basis for methylphosphonate biosynthesis. Science. 358(6368):1336-1339. PMID: 29217579
Ju, K.S., X. Xhang, and M.A. Elliot. 2017. New kid on the block: LmbU expands the repertoire of specialized metabolic regulators in Streptomyces. J. Bacteriol. 200(2):pii:200559-17. PMID: 29084856
Freestone, T.F., K.S. Ju, B. Wang, and H. Zhao. 2017. Discovery of a phosphonoacetic acid derived natural product by pathway refactoring. ACS Synth. Biol. 6(2):217-223. PMID: 28103011
Labeda, D.P., C.A. Dunlap, X. Rong, Y. Huang, J.R. Doroghazi, K.S. Ju, and Metcalf, W.W. 2017. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 110(4):563-583. PMID: 28039547
Amato, K., A. Ulanov, K.S. Ju, and P. Garber. 2017. Metabolomic data suggest regulation of black howler monkey (Alouatta pigra) diet composition at the molecular level. Am. J. Primatol. 79(4):1-10. PMID: 27936282
McClure, R.A.; Goering, K.S. Ju, J. Baccile, F.C. Shroeder, W.W. Metcalf, R.J. Thomson, and N.L. Kelleher. 2016. Elucidating the rimosamide-detoxin natural product families and their biosynthesis using metabolite/gene cluster correlations. ACS Chem. Biol. 11(12): 3452-3460. PMID: 27809474
Goering, A.W., R.A. McClure, J.R. Doroghazi, J.C. Albright, N.A. Haverland, Y. Zhang, K.-S. Ju, R.J. Thomson, W.W. Metcalf, and N.L. Kelleher. 2016. Metabologenomics: correlation of microbial genes with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Central Science. 2:99-108. PMID: 27163034
Labeda, D.P., X. Rong, Y. Huang, J.R. Doroghazi, K.-S. Ju, and W.W. Metcalf. 2016. Taxonomic evaluation of species in the Streptomyces hirsutus clade using multi-locus sequence analysis and proposals to reclassify several species in this clade. International Journal of Systematic and Evolutionary Microbiology. 66:2444-2450. PMID: 26971011
Ju, K.-S., J. Gao, J.R. Doroghazi, K.A. Wang, C.J. Thibodeaux, S. Li, E. Metzger, J. Fudala, J. Su, J. Zhang, J.P. Cioni, J. Lee, B.S. Evans, R. Hirota, D.P. Labeda, W.A. van der Donk, and W.W. Metcalf. 2015. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proceedings of the National Academy of Sciences USA. 112:12175-12180. PMID: 26324907
Mahan, K.M., J.T. Penrod, K.-S. Ju, N. Al Kass, W.A. Tan, R. Troung, J.V. Parales, and R.E. Parales. 2015. Selection of growth on 3-nitrotoluene identifies nitroarene dioxygenases with altered specificities. Applied and Environmental Microbiology. 81:309-319. PMID: 25344236
Doroghazi, J.R.*, J.C. Albright*, A.W. Goering, K.-S. Ju, R.R. Haines, K.A. Tchalukov, D.P. Labeda, N.L. Kelleher, and W.W. Metcalf. 2014. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nature Chemical Biology. 10:963-968. PMID: 25262415
Ju, K.-S., J.R. Doroghazi, and W.W. Metcalf. 2014. Genomics-enabled discovery of phosphonate natural products and their biosynthetic pathways. Journal of Industrial Microbiology and Biotechnology. 41:345-356. PMID: 24271089
Gao, J., K.-S. Ju, X. Yu, J.E. Velasquez, S. Mukherjee, J. Lee, C. Zhao, B.S. Evans, J.R. Doroghazi, W.W. Metcalf, and W.A. van der Donk. 2014. Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster. Agewandte Chem Int Ed Engl. 53:1334-1337. PMID: 24376039
Cioni, J.C, J.R. Doroghazi, K.-S. Ju, X. Yu, B.S. Evans, and W.W. Metcalf. 2014. A cyanohydrin phosphonate natural product from Streptomyces regensis. Journal of Natural Products. 77:243-249. PMID: 24437999
Labeda, D.P., J.R. Doroghazi, K.-S. Ju, and W.W. Metcalf. 2014. Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidicus comb. nov. International Journal of Systematic and Evolutionary Microbiology. 64:894-900. PMID: 24277863
Evans, B.S., C. Zhao, J. Gao, C.M. Evans, K.-S. Ju, J.R. Doroghazi, W.A. van der Donk, N.L. Kelleher, and W.W. Metcalf. 2013. Discovery of the antibiotic phosacetamycin via new mass spectrometry-based method for phosphonic acid detection. ACS Chemical Biology. 8:908-913. PMID: 23474169
