
Mingxu Fang
Assistant Professor of Microbiology
428A Biological Sciences Building
484 W. 12th Ave
Areas of Expertise
- Microbial Physiology
- Gene Regulation
- Circadian Biology
Education
- Postdoc, University of California, San Diego, 2018-2024
- Ph.D. Indiana University, Bloomington, 2018
- B.S., Zhejiang University, 2012
Research
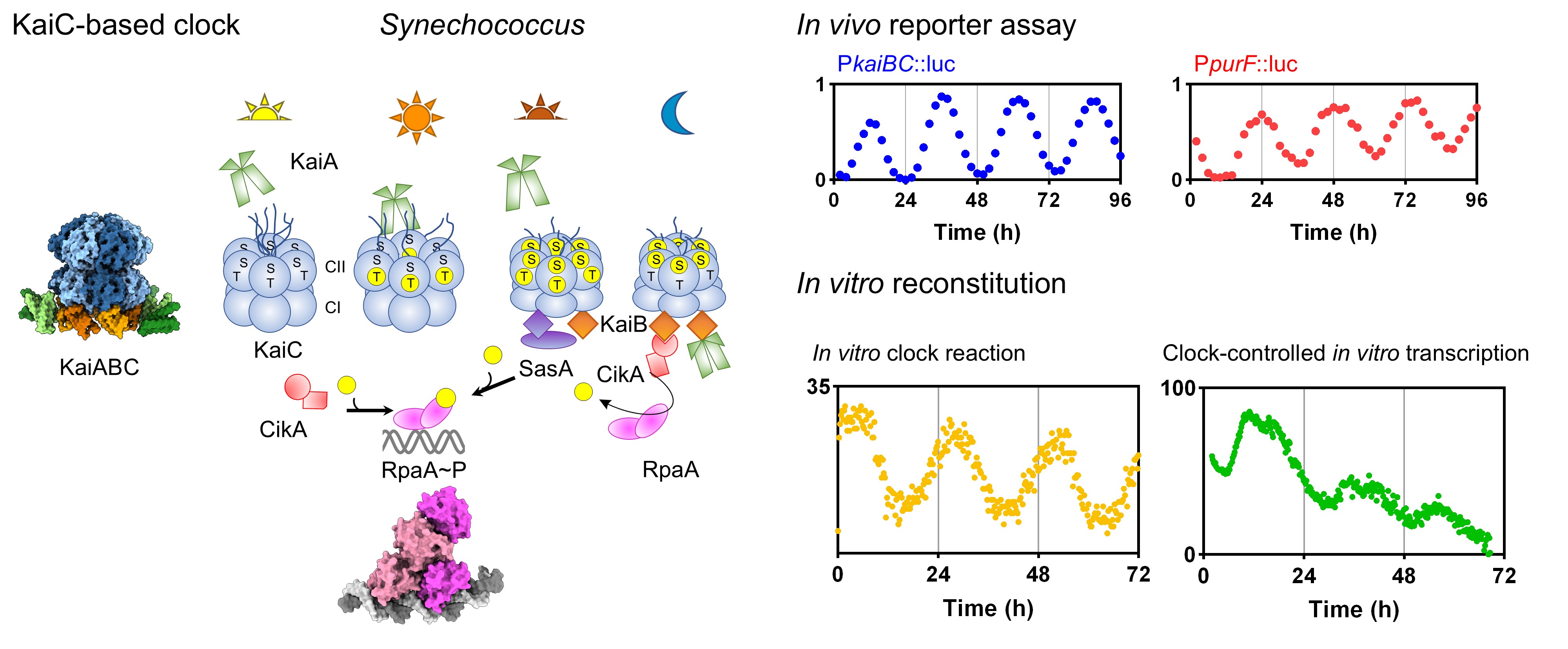
The general interest of the Fang lab lies in understanding how the environment shapes the physiology of microorganisms at the molecular level. Currently, we focus on the circadian clock in the cyanobacterium Synechococcus elongatus. Like all life forms on Earth, bacteria are also subjected to environmental changes associated with the day-night cycle. Over evolutionary time, these predictable cycles have exerted selective pressure, driving the development of internal timing mechanisms that anticipate recurrent environmental changes. By aligning the endogenous rhythm of cellular processes with the 24-h environmental cycle, the S. elongatus circadian clock confers fitness to cells.
The main driver of the S. elongatus circadian rhythm is an oscillator composed of interactions among three proteins: KaiA, KaiB, and KaiC. This oscillator regulates gene expression through an output pathway, ensuring that specific genes turn on or off at precise times of the day. We use a combination of genetic, biochemical, bioinformatic, and structural approaches to uncover the molecular mechanisms underlying this regulation. The extensive genetic tools available for S. elongatus enable a top-down approach to identify and modify clock components in vivo. Additionally, a remarkable feature of the KaiABC oscillator—its ability to be reconstituted in vitro—offers a unique bottom-up approach to dissect biological clock mechanisms in unparalleled detail. More recently, we have leveraged high-throughput real-time biochemical assays to recapitulate clock-controlled gene transcription in vitro.
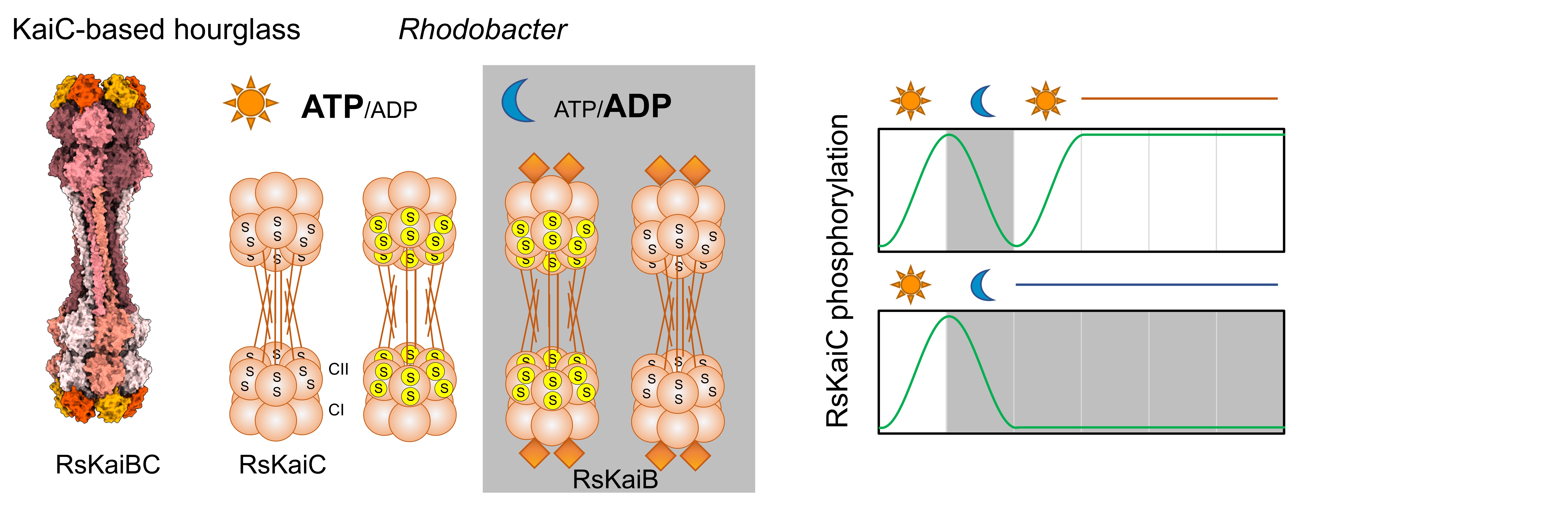
Another research direction in the lab is exploring alternative timing mechanisms in bacteria. KaiC homologs have been identified in various bacterial phyla, and recent structural and biochemical studies on KaiBC from purple non-sulfur bacteria Rhodobacter sphaeroides suggest that this system functions as an hourglass rather than an autonomous clock, requiring external resets to maintain timing. We are particularly interested in understanding how and why R. sphaeroides uses this hourglass mechanism to regulate its physiology.
Selected Publications
Fang M, LiWang A, Golden SS, Partch, C. (2024) The inner workings of the tiniest biological clock. Trends Biochem Sci 49(3):236-246
Fang M, Chavan AG, LiWang A, Golden SS. (2023) Synchronization of the circadian clock to the environment tracked in real time. Proc Natl Acad Sci U S A 120: e2221453120. (This work was highlighted in a companion commentary in the same issue by Larrondo LF; Proc Natl Acad Sci U S A 120: e2303566120)
Weiss EL, Fang M, Taton A, Szubin R, Palsson BØ, Mitchell BG, et al. (2022) An unexpected role for leucyl aminopeptidase in UV tolerance revealed by a genome-wide fitness assessment in a model cyanobacterium. Proc Natl Acad Sci U S A 119: e2211789119.
Chavan AG, Swan A, Heisler J, Sancar C, Ernst DC, Fang M, et al. (2021) Reconstitution of an intact clock reveals mechanisms of circadian timekeeping. Science 374(6564).
Fang M, Bauer CE. (2018) Regulation of stringent factor by branched chain amino acids. Proc Natl Acad Sci U S A 115: 6446-6451. (This work was recommended by F1000)
