Brian Ahmer
Contact Information
Professor of Microbial Infection & Immunity
Areas of Expertise
- Detection of microbes and host environment by Salmonella
Education
- B.S. Colorado State University, 1990
- Ph.D. Washington State University, 1994
- Postdoc, Oregon Health Sciences University, 1994-1999
Research Interests
We are studying how Salmonella thrives in the host environment and how to treat bacterial infections. We are currently focusing on three projects:
Detection of other microbes by Salmonella
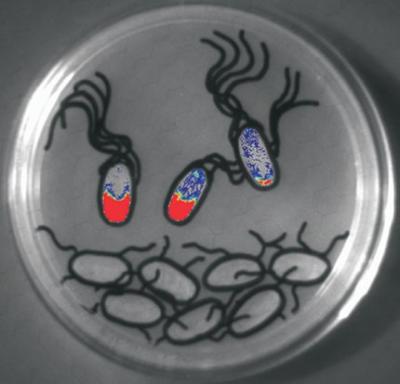
Some bacteria use pheromones (N-acylhomoserine lactones or AHLs) to determine their population density. Salmonella does not make AHLs but it can detect the AHLs produced by other bacteria. We hypothesized that Salmonella would detect AHLs made by the normal intestinal microbiota and use this information to adjust its gene expression accordingly. Surprisingly, we discovered that the normal gut microbiota does not make AHLs and instead, Salmonella is detecting the AHL production of other pathogens in the gut. Salmonella can detect the AHL production of Aeromonas hydrophila in turtles and Yersinia enterocolitica in mice. The picture to the left is a photo of an LB agar plate upon which Salmonella encoding a pheromone-responsive luciferase fusion is struck in the top three cartoon bacteria, while the pheromone-producing organism, Hafnia alvei, is struck in the cartoon bacteria below. The Salmonella nearest the Hafnia are producing light.
A Multi-omics Approach to Studying Infection
In collaboration with the Kelly Wrighton and Vicki Wysocki labs we are characterizing Salmonella infection of the mouse using several omics technologies. We are characterizing the effect of Salmonella on the intestinal microbial community using 16S profiling and shotgun metagenomics; we are characterizing the gene expression of Salmonella and the rest of the community using metatranscriptomics; and the metabolites in the system using metabolomics. This is done at multiple time points to obtain a dynamic view of the infection. Integration of this data will generate large numbers of testable hypotheses. Future projects include testing these hypotheses by constructing Salmonella mutants to see how they behave in various mouse models, including gnotobiotic mouse models (mice with a defined microbial community composition).
Killing Bacteria by Inducing Sugar-Phosphate Toxicities
We discovered that fructose-asparagine is a nutrient for Salmonella in the inflamed intestine. While characterizing the enzymology and regulation of this system, we determined that inhibition of the FraB enzyme in the pathway causes the accumulation of a metabolite that is toxic to Salmonella. This makes FraB an interesting drug target. Since then, we have expanded to look at many more metabolic pathways that include toxic intermediates. Our current favorite is the mannitol utilization pathway. Since mannitol is used in human medicine, and reaches bacteria at systemic sites, inhibitors of MtlD in an IV or beverage containing mannitol could be a new method of treating infections. We are actively screening compounds to find inhibitors of MtlD. Future projects include determining why mannitol is toxic to a Salmonella mtlD mutant (what is the mechanism of intoxication?) and determining how Salmonella recovers from these effects and how Salmonella might become resistant to MtlD inhibitors.
Relevant Publications
- Sabag-Daigle A, Blunk HM, Sengupta A, Wu J, Bogard AJ, Ali MM, et al. A metabolic intermediate of the fructose-asparagine utilization pathway inhibits growth of a Salmonella fraB mutant. Sci Rep. 2016;6:28117; doi: 10.1038/srep28117.
- Sabag-Daigle A, Wu J, Borton MA, Sengupta A, Gopalan V, Wrighton KC, et al. Identification of bacterial species that can utilize fructose-asparagine. Appl Environ Microbiol. 2018;84(5); doi: 10.1128/AEM.01957-17.
- Wu J, Sabag-Daigle A, Metz TO, Deatherage Kaiser BL, Gopalan V, Behrman EJ, et al. Measurement of fructose-asparagine concentrations in human and animal foods. J Agric Food Chem. 2018;66(1):212-7; doi: 10.1021/acs.jafc.7b04237.
- Boulanger EF, Sabag-Daigle A, Thirugnanasambantham P, Gopalan V, Ahmer BMM. Sugar-phosphate toxicities. Microbiol Mol Biol Rev. 2021:e0012321; doi: 10.1128/MMBR.00123-21.
- Boulanger EF, Sabag-Daigle A, Baniasad M, Kokkinias K, Schwieters A, Wrighton KC, et al. Sugar-phosphate toxicities attenuate Salmonella fitness in the gut. J Bacteriol. 2022;204(12):e0034422; doi: 10.1128/jb.00344-22.
- Sabag-Daigle A, Boulanger EF, Thirugnanasambantham P, Law JD, Bogard AJ, Behrman EJ, et al. Identification of small-molecule inhibitors of the Salmonella FraB deglycase using a live-cell assay. Microbiol Spectr. 2023;11(2):e0460622; doi: 10.1128/spectrum.04606-22.
- Kokkinias K, Sabag-Daigle A, Kim Y, Leleiwi I, Shaffer M, Kevorkian R, et al. Time-resolved multi-omics reveals diverse metabolic strategies of Salmonella during diet-induced inflammation. Msphere. 2024:e0053424; doi: 10.1128/msphere.00534-24.
- Leleiwi I, Kokkinias K, Kim Y, Baniasad M, Shaffer M, Sabag-Daigle A, et al. Gut microbiota carbon and sulfur metabolisms support Salmonella infections. Isme j. 2024;18(1); doi: 10.1093/ismejo/wrae187.
- Schwieters A, Ahmer BMM. Identification of new SdiA regulon members of Escherichia coli, Enterobacter cloacae, and Salmonella enterica serovars Typhimurium and Typhi. Microbiol Spectr. 2024:e0192924; doi: 10.1128/spectrum.01929-24.
- Schwieters A, Cole AL, Rego E, Gao C, Kebriaei R, Wysocki VH, et al. MtlD as a therapeutic target for intestinal and systemic bacterial infections. J Bacteriol. 2024:e0048024; doi: 10.1128/jb.00480-24.
The all-time favorite:
Schwieters A, Cole AL, Rego E, Gao C, Kebriaei R, Wysocki VH, et al. MtlD as a therapeutic target for intestinal and systemic bacterial infections. J Bacteriol. 2024:e0048024; doi: 10.1128/jb.00480-24.
