Areas of Expertise
- Molecular biology of archaea
- Molecular adaptations to extreme environments
Education
- B.S. University of Birmingham, UK, 1968
- Ph.D. University of British Columbia, 1971
- Postdoc, University of Arizona, 1971-1973
- Postdoc, Max Planck Institute, W. Berlin, 1974-1979
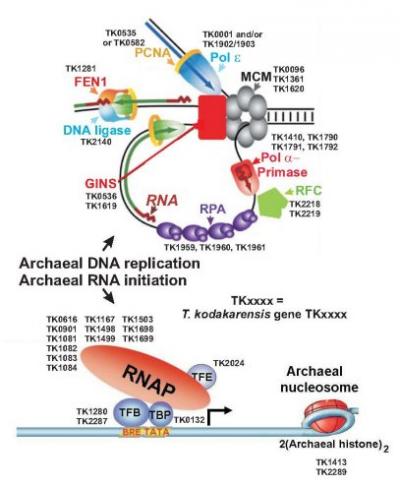
Research Interests
Archaeal gene expression
Archaea are prokaryotes, many of which have very unusual life-styles and unique biochemical abilities. Our research goal is to define and understand the molecular machinery and regulatory systems involved in gene expression in two thermophilic Euryarchaea, Thermococcus kodakarensis (T.k.) and Methanothermobacter thermautotrophicus (M.t).
Archaeal bio-energy production
T.k. is an anaerobic heterotroph that grows optimally at 85ºC by digesting starch, chitin or peptides and produces hydrogen. M.t.is an anaerobic autotroph that grows optimally at 65ºC by consuming carbon dioxide and hydrogen and so produces methane. Our experiments combine genetics and biochemistry to identify the molecular components, catalysts and regulation of these globally and commercially very important archaeal bio-energetic systems.
Archaeal histones and chromatin
Relevant Publications
-
Nalabothula N, Xi L, Bhattacharyya S, Widom J, Wang J-P, Reeve JN, Santangelo TJ & Fondufe-Mittendorf YN (2013) Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs.BMC Genomics 14:391.
-
Čuboňová L, Richardson T, Burkhart BW, Kelman Z, Connoll BA, Reeve JN & Santangelo, TJ (2013) Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J Bacteriol 195:2322-8.
-
Pan M, Santangelo TJ, Čuboňová L, Li Z, Metangmo H, Ladner J, Hurwitz J, Reeve JN & Kelman Z (2013) Thermococcus kodakarensis has two functional PCNA homologs but only one is required for viability. Extremophiles 17:453-61.
-
Čuboňová L, Katano M, Kanai T, Atomi H, Reeve JN & Santangelo TJ (2012) An archaeal histone is required for transformation of Thermococcus kodakarensis. J Bacteriol 194:6864-74.
-
Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN & Kelman Z (2011) A novel DNA nuclease is stimulated by association with the GINS complex. Nucl Acids Res 39:6114-23.
-
Santangelo TJ, Čuboňová L & Reeve JN (2011) Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol Microbiol 81:897-911.
-
Li Z, Santangelo TJ, Cuboňová L, Reeve JN & Kelman Z (2010) Affinity purification of an archaeal DNA replication protein network.Mbio 1: e00221-10.
-
Santangelo TJ, Čuboňová L & Reeve JN (2010) Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol 74:1044-52.
-
Santangelo TJ & Reeve JN (2010) Deletion of switch 3 results in an archaeal RNA polymerase that is defective in transcript elongation. J Biol Chem 285:23908-15.
-
Jahn U, Aigner J, Längst G, Reeve JN & HuberH (2009) Nanoarchaeal origin of histone H3? J Bacteriol 191:1092-6.
Historical favorites:
- French SL, Santangelo TJ, Beyer AL & Reeve JN. (2007) Transcription and translation are coupled in Archaea. Mol Biol Evol. 24, 893-5.
- Sandman K, Krzycki JA, Dobrinski B, Lurz R & Reeve JN. (1990) DNA binding protein HMf from the hyperthermophilic archaebacterium Methanothermus fervidus is most closely related to histones. Proc Natl Acad Sci U S A 87, 5788-91.
