Areas of Expertise
- Phenotypic consequences of genomic variation
Education
- Ph.D. Stanford University, 2009
Research Interests
Our research focus is on the evolution of genes and genomes in the major human fungal pathogen Candida albicans and how these genetic changes contribute to phenotypic diversity. C. albicans resides as a harmless commensal in the majority of people worldwide but is capable of causing superficial mucosal infections in otherwise healthy individuals as well as life-threatening systemic infections in patients with compromised immune systems.
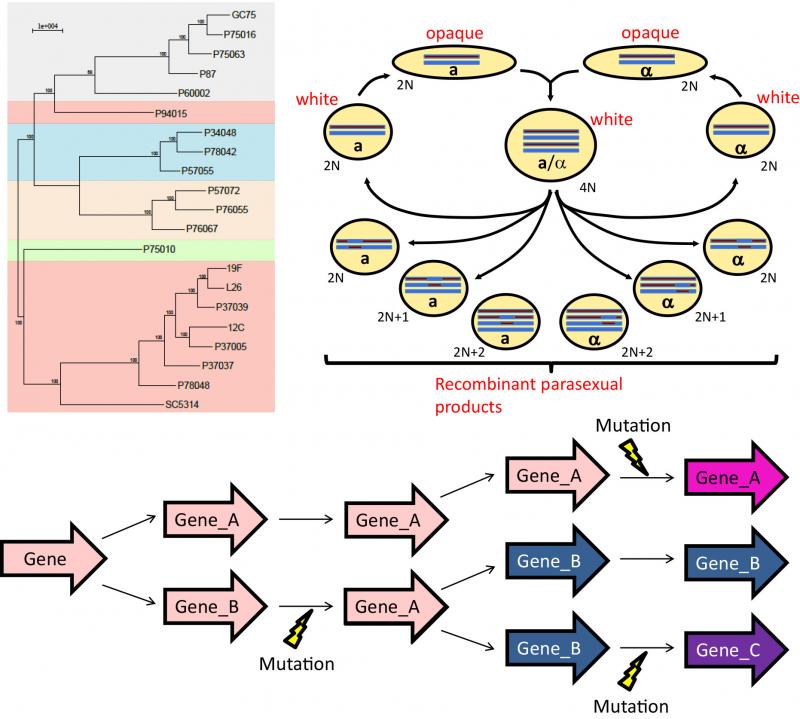
Genetic basis of phenotypic diversity
Widespread phenotypic diversity is found among C. albicans clinical isolates, including variability in the extent of disease produced in insect and animal models of infection. This variation is produced by massive genetic diversity between C. albicans strains, which is similar in magnitude to the nucleotide divergence between humans and chimpanzees. We are working to identify the genetic basis for C. albicans phenotypic diversity and explore how individual polymorphisms affect host/pathogen interactions using combined quantitative genetic approaches. Differential host response to C. albicans isolates remains an underexplored topic and our research will provide insight into how natural selection acts upon this variation in the context of commensalism and infection.
Expanded gene families in virulence
The telomere-associated (TLO) gene family has expanded significantly in C. albicans relative to less pathogenic Candida species. These genes encode components of the Mediator complex, a key transcriptional regulator complex in eukaryotes. Significant strain-to-strain variation in gene copy number exists for TLOs although the role of individual paralogs is unclear. We are investigating the molecular and biological roles of individual TLO genes to understand what selective pressures exist to promote gene family expansion and evolution.
The role of eukaryotes in the human gut microbiome
Microbial eukaryotes are an integral component of human-associated microbiota but are often neglected in understanding the ecological framework of these host niches and in contributions towards health and disease. Species of fungi, protists, and helminths have all been identified within the human gut but there relative abundance across different populations and conditions remains unclear. We have recently developed a metagenomics approach to identify eukaryotes present at low abundances in complex microbial communities and are working to understand their role in community architecture in these niches and their importance towards human health.
Relevant Publications
Dunn, M.J., Shazib, S.U.A., Simonton, E., Slot, J., and Anderson, M.Z. Architectural groups of a subtelomeric gene family evolve along distinct paths in Candida albicans (G3, in press).
Nastasi, N., Anderson, M. Z., Balasubrahmaniam, N., Bope, A., Britt Jr., R. D., Cochran, S. J., Gill, Hegarty, B., Lee, J., Stamps, B. W., Stickney, A., Taylor, C. G., Tram, N. K., Van Dusen. J., Varaljay, V. A., Weir, M. H., Horack, J. M., Oglesbee, M., Sullivan, M. B., and Dannemiller, K. C., “Application of emerging innovations in microbiome science to space development and settlement systems,” 73rdInternational Astronautical Congress, Paris, France, September 18-22, 2022.
Sindledecker, D., Dunn, M.J., Anderson, M.Z., and Stoodley, P. 2022 Genomic and transcriptomic profiling of phoenix colonies. Sci Reports. 12: 13726. PMID: 35962051.
McCartney, A.M., Anderson, J., Liggins, L., Hudson, M., Anderson, M.Z., TeAika, B., Geary, J., Cook-Deegan, R., Patel, H., and Phillippy, A.M. 2022. Balancing openness with Indigenous Data Sovereignty – an opportunity to leave no one behind in the journey to sequence all of life. PNAS. 119: e2115860119.
Mishra, A., Forche, A., and Anderson, M.Z. 2021. Parasex in Candida albicans. Frontiers Cell Infect Micro. 11: 796929. PMID: 34966696.
Wang, J.M.*, Woodruff, A.L.*, Dunn, M.J., Fillinger, R.J., Bennett, R.J., and Anderson, M.Z. 2021. Intraspecies transcriptional profiling reveals key regulators of Candida albicans pathogenic traits. mBio. 12: e00586-21. PMID 33879584.
Serrano, J., Smith, K.R., Crouch, A.L., Sharma, V., Yi, F., Vargova, V., LaMoia, T.E., Dupont, L.M., Serna, V., Tang, F., Gomes-Dias, L., Blakeslee, J., Hatzakis, E., Peterson, S.N., Anderson, M., Pratley, R.E., and Kyriazis, G.A. 2021. High-dose saccharin supplementation does not induce gut microbiota changes or glucose intolerance in healthy humans and mice. Microbiome. 12: 11. PMID 33431052
Dunn, M.J., Fillinger, R.J., Anderson, L.M., and Anderson, M.Z. 2020. Automated quantification of biofilm-related phenotypes reveals additive contribution to biofilm production. npj Biofilms & Microbiomes. 6: 36. PMID 33037223
Sindledecker, D., Moore, K., Li, A., Wozniak, D.J., Anderson, M.Z., Dusane, D.H., Stoodley, P. 2020. Novel aminoglycoside tolerant “phoenix” colony variants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 64: e00623-20. PMID 32540981
Chemudupati, M., Smith, A., Fillinger, R.J., Kenney, A., Zhang, L., Zani, A., Liu, S.L., Anderson, M.Z., Sharma, A., and Yount, J. 2020. Butyrate reprograms expression of specific interferon stimulated genes. J. Virol. 94: e00326-20. PMID 32461320
Cullen, C.M., Beyhan, S., Cho, C.E., Wolosyznek, S., Convertino, M., McCoy, S.J., Zhang, Y., Anderson, M.Z., Alvarez-Ponce, D., Smirnova, E., Karstens, L., Dorrestein, P.C., Li, H., Gupta, A.S., Cheung, K., Powers, J.G., Zhao, Z., Rosen, G. 2020. Emerging priorities for microbiome research. Frontiers in Microbiology. 11: 136.
Dunn, M.J. and Anderson M.Z. 2019. To repeat or not to repeat: Repetitive sequences regulate genome stability in Candida albicans. Genes. 10: e866.
Ene, I.V., Bennett, R.J., and Anderson, M.Z. 2019. Mechanisms of genome evolution in Candida albicans. Curr Op Micro. 52: 47-54.
Anderson, M.Z.*, Thompson, G., Clark, M., Hirakawa, M.P., and Bennett, R.J.* 2019. A ‘parameiosis’ drives depolyploidization and homologous recombination in Candida albicans. Nat Comm. 10: 4388.
Anderson, M.Z. 2019. Considering Complex Mutational Processes that Shape Microbial Virulence. mSphere. 4: e00578-19.
Fillinger, R.J. and Anderson, M.Z. 2019. Seasons of change: Genome evolution of human fungal pathogens. Infection, Genetics and Evolution. 70: 165-174.
Liang, S., Anderson, M.Z., Hirakawa, M.P., Wang, J.M., Ene, I.V., Frazer, C., Alaalm, L.M., Thomson, G.J., and Bennett, R.J. 2019. Hemizygosity enables a mutational transition governing fungal virulence and commensalism. Cell Host & Microbe. 25: 1-14.
Monsey, L., Best, L.G., Zhu, J., DeCroo, S., and Anderson, M.Z. 2019. The Association of Mannose Binding Lectin Genotype and Immune Response to Chlamydia pneumoniae: The Strong Heart Study. PLoS One. 14: e0210640.
Moran, G.C., Anderson, M.Z., Myers, L.C., and Sullivan, D.J. 2019. Role of Mediator in virulence and antifungal drug resistance in pathogenic fungi. Curr Genet. 65:1-10.
Dunn, M.J., Woodruff, A., and Anderson, M.Z. 2018. The Galleria mellonella waxworm infectious model for disseminated candidiasis. J. Vis. Exp. 141: e58914.
Wang, J.A., Bennett, R.J., and Anderson, M.Z. 2018. The genome of the human pathogen Candida albicans is shaped by mutation and cryptic sexual recombination. mBio. 9: e01205-18.
Uppuluri, P., Liu, N.N., Dunn, M.J., Anderson, M.Z., Berman, J., Lopez-Ribot, J.L., and Koehler, J.. 2018. Transcriptome characteristics of Candida albicans biofilm dispersed cells sheds light on the regulation of the biofilm dispersal process. mBio. 9: e01338-18.
Claw, K.G., Anderson, M.Z., Begay, R.L., Tsosie, K.S., Fox, K., Garrison, N.A., the SING Consortium. 2018. Shifting the Research Paradigm: Indigenous Genomics and Health. Nat Comm. 9: 2957. PMID: 30054469.
Dunn, M.J., Kinney, G.M., Washington, P.M., Berman, J., Anderson, M.Z.. Functional diversification accompanies gene family expansion of MED2 homologs in Candida albicans. PLoS Genet 14(4): e1007326. https://doi.org/10.1371/journal.pgen.1007326
Scaduto, C.M., Kabrawala, S., Scheving, W., Ly, A., Anderson, M.Z., Whiteway, M., and Bennett, R.J.. Epigenetic control of pheromone MAPK signaling determines sexual fecundity in Candida albicans. PNAS. Dec 2017, 114 (52) 13780-13785; DOI: 10.1073/pnas.1711141115. PMID: 29255038.
Kenny, A., Dowdle, J.A., Bozzacco, L., McMichael, T.M., St Gelais, C., Panfil, A.R., Sun, Y., Anderson, M.Z., Green, P.L., Lopez, C.B., Rosenberg, B.R., Wu, L., and Yount, J.S. 2017. Human genetic determinants of viral diseases. Annual Rev Genetics. 51: 241-263. PMID 28853921.
Anderson, M.Z., Saha, A., Haseeb, A., and Bennett, R.J. 2017. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinincal isolate of Candida albicans. Microbiology. 163: e000478. PMID 28640746.
Anderson, M.Z., Porman, A.P., Wang, N., Huang, D., Cuomo, C., and Bennett, R.J. 2016. A multistate toggle switch defines fungal cell fates and is regulated by synergistic genetic cues. PLoS Genetics. 12: e1006353.
Anderson, M.Z. and Bennett, R.J. 2016. Budding off: Bringing functional genomics to Candida albicans. Briefings in Functional Genomics. 15: 85-94.
Anderson, M.Z., Wigen, L., Burrack, L.S., and Berman, J. 2015. Real-time gene movement propels subtelomeric evolution in Candida albicans. Genetics. 200: 907-19.
Hirakawa, M.P., Martinez, D.A.*, Sakthikumar, S.*, Anderson, M.Z., Berlin, A., Gujja, S., Zeng, Q., Zisson, E., Wang, J.A., Greenberg, J.A., Berman, J., Bennett, R.J., and Cuomo, C.A. 2015. Intra-species variation in Candida albicans: Aneuploidy, loss of heterozygosity, and balancing commensalism and pathogenesis. Genome Research. 25: 413-25.
Haran, J., Boyle, H., Hokamp, K., Yeomans, T., Liu, Z., Church, M., Fleming, A., Anderson, M.Z., Berman, J., Myers, L.C., Sullivan, D.J., and Moran G. 2014. Telomeric ORFs (TLOs) in Candida spp. encode Mediator subunits that regulate distinct virulence traits. PLoS Genetics. 10: e1004658.
Anderson, M.Z., Gerstein, A.C., Wigen, L., Baller, J.A., and Berman, J. 2014. Silencing is noisy: Population and cell level noise in telomere-adjacent genes is dependent on telomere position and Sir2. PLoS Genetics. 10: e1004436.
Anderson, M.Z., Baller, J.A., Dulmage, K., Wigen, L., and Berman, J. 2012. The three clades of the telomere-associated TLO gene family of Candida albicans have different splicing, localization and expression features. Eukaryot Cell. 11:1268-75.
Anderson, M.Z., Brewer, J.L., Singh, U., and Boothroyd, J.C. 2009. A Pseudouridine Synthase Homologue Is Central to Cellular Differentiation in Toxoplasma gondii. Eukaryotic Cell. 3:398-409.

